A tweet from @coneee yesterday about merging two datasets using columns of data that don’t quite match got me wondering about a possible R recipe for handling partial matching. The data in question related to country names in a datafile that needed fusing with country names in a listing of ISO country codes,although the recipe I’m describing here is intended to be a general purpose partial matcher. (See the comments for alternative methods for the special case of reconciling country names with country codes.)
The original data set had the form:
RANK,ECONOMY,PERCENTAGE OF INDIVIDUALS USING THE INTERNET 2011 1,Iceland,95 2,Norway,94 3,Netherlands,92.3 4,Sweden,91 5,Luxembourg,90.9 6,Denmark,90 7,Finland,89.4 8,Qatar,86.2 9,New Zealand,86 10,Switzerland,85.2 11,Liechtenstein,85 12,S. Korea,83.8 13,Germany,83 14,Canada,83 15,United Kingdom,82 16,Antigua & Barbuda,82
I’m not sure what country code listing was being used, but it probably looked something like this list of ISO Country Codes by Country:
ANDORRA;AD ANGOLA;AO ANGUILLA;AI ANTARCTICA;AQ ANTIGUA AND BARBUDA;AG ARGENTINA;AR ARMENIA;AM ARUBA;AW AUSTRALIA;AU AUSTRIA;AT AZERBAIJAN;AZ BAHAMAS;BS BAHRAIN;BH
It’s easy enough to reduce all the country names to lowercase characters so that we can try to match them exactly without worrying about any differences in capitalisation, but how do we match country names that don’t match exactly – ANTIGUA AND BARBUDA and Antigua & Barbuda, for example, or Central African Rep. and Central African Republic?
One trick is to use one of the well known partial string matching algorithms, such as the Levenshtein Distance. Here’s a recipe I hacked together that first tries to find an exact match on country names by attempting to merge the two country lists directly, and then tries to partially match any remaining unmatched names in the original list. A signature function is constructed to help out the partial matching attempt that reduces all words in the country name to lower case, sorts them alphabetically, and then concatenates them with no adjoining spaces.
(The signature idea was cribbed from the fingerprint that is available in Google Refine and that I employed in Merging Datasets with Common Columns in Google Refine.)
[@downes commented that the code wasn’t really written with clarity in mind – so I’ve added some comments…]
#Load in the data from the URLs:
PercentageUsingTheNet=read.csv(url('http://s.telegraph.co.uk/graphics/conrad/PercentageUsingTheNet.csv, encoding='MACROMAN'))
ccode=read.csv(url('http://www.iso.org/iso/country_names_and_code_elements_txt'),sep=';')
##Here's where the algorithm starts...
##I'm going to generate a signature from country names to reduce some of the minor differences between strings
##In this case:
### convert all characters to lower case (tolower())
### split the string into a vector (unlist()) of separate words (strsplit())
### sort the words alphabetically (sort())
### and then concatenate them with no spaces (paste(y,collapse='')).
##So for example, United Kingdom would become kingdomunited
##To extend this function, we might also remove stopwords such as 'the' and 'of', for example (not shown).
signature=function(x){
sig=paste(sort(unlist(strsplit(tolower(x)," "))),collapse='')
return(sig)
}
#The partialMatch function takes two wordlists as vectors (x,y) and an optional distance threshold (levDist)
#The aim is to find words in the second list (y) that match or partially match words in the first (x)
partialMatch=function(x,y,levDist=0.1){
#Create a data framecontainind the signature for each word
xx=data.frame(sig=sapply(x, signature),row.names=NULL)
yy=data.frame(sig=sapply(y, signature),row.names=NULL)
#Add the original words to the data frame too...
xx$raw=x
yy$raw=y
#We only want words that have a signature...
xx=subset(xx,subset=(sig!=''))
#The first matching pass - are there any rows in the two lists that have exactly the same signature?
xy=merge(xx,yy,by='sig',all=T)
matched=subset(xy,subset=(!(is.na(raw.x)) & !(is.na(raw.y))))
#?I think matched=xy[ complete.cases(raw.x,raw.y) ] might also work here?
#Label the items with identical signatures as being 'Duplicate' matches
matched$pass="Duplicate"
#Grab the rows from the first list that were unmatched - that is, no matching item from the second list appears
todo=subset(xy,subset=(is.na(raw.y)),select=c(sig,raw.x))
#We've grabbed the signature and original raw text from the first list that haven't been matched up yet
#Name the columns so we know what's what
colnames(todo)=c('sig','raw')
#This is the partial matching magic - agrep finds items in the second list that are within a
## certain Levenshtein distance of items in the first list.
##Note that I'm finding the distance between signatures.
todo$partials= as.character(sapply(todo$sig, agrep, yy$sig,max.distance = levDist,value=T))
#Bring the original text into the partial match list based on the sig key.
todo=merge(todo,yy,by.x='partials',by.y='sig')
#Find the items that were actually partially matched, and pull out the columns relating to signatures and raw text
partial.matched=subset(todo,subset=(!(is.na(raw.x)) & !(is.na(raw.y))),select=c("sig","raw.x","raw.y"))
#Label these rows as partial match items
partial.matched$pass="Partial"
#Add the set of partially matched items to the set of duplicate matched items
matched=rbind(matched,partial.matched)
#Find the rows that still haven't been matched
un.matched=subset(todo,subset=(is.na(raw.x)),select=c("sig","raw.x","raw.y"))
#If there are any unmatched rows, add them to the list of matched rows, but labelled as such
if (nrow(un.matched)>0){
un.matched$pass="Unmatched"
matched=rbind(matched,un.matched)
}
#Grab the columns of raw text from x and y from the matched list, along with how they were matched/are unmatched
matched=subset(matched,select=c("raw.x","raw.y","pass"))
#Ideally, the length of this should be the same as the length of valid rows in the original first list (x)
return(matched)
}
#A rogue character in @coneee's data file borked things for me, so I originally needed to do a character code conversion first
#PercentageUsingTheNet$ECONOMY=iconv(PercentageUsingTheNet$ECONOMY)
#Loading the CSV with the encoding attribute set (h/t Kent Johnson) seemed to work properly though...
#Call the partial match function using two vectors
#The aim is to find items in the second vector that partially match items in the first
#The function returns the first list annotated with partial match items from the second and a match type
matches=partialMatch(PercentageUsingTheNet$ECONOMY,ccode$Country.Name)
As ever, this code was arrived at by tinkering and searching on Stack Overflow (using search terms along the lines of “partial match R” and “R levenshtein”). If you can improve on it, please do so and paste a link to the improved code, or a code fragment itself, in the comments below:-)
UPDATE: via the comments, the following suggestion that I don’t have time to check right now…
#Bring the original text into the partial match list based on the sig key.
-todo=merge(todo,yy,by.x=’partials’,by.y=’sig’)
+todo=merge(todo,yy,by.x=’partials’,by.y=’sig’,all.x=T)
#Label these rows as partial match items
-partial.matched$pass=”Partial”
+if (nrow(partial.matched) > 0) partial.matched$pass=”Partial”
Thanks:-)
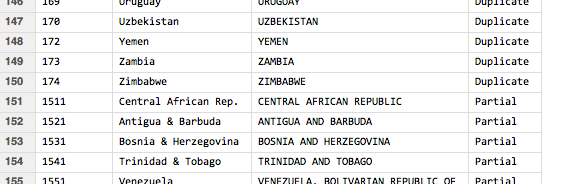
When we run the script and look at the contents of the matches dataframe, this is an example of what we get:
This data frame can then be merged with the originally loaded data to give us the required country code annotations:
a=PercentageUsingTheNet
b=ccode
#Merge the original data set with the ISO country code country name keys
aa=merge(a,matches,by.x='ECONOMY',by.y='raw.x',all.x=T)
#Merge in the ISO country codes
aa=merge(aa,b,by.x='raw.y',by.y='Country.Name',all.x=T)
aa=subset(aa,select=c('ECONOMY','RANK','PERCENTAGE.OF.INDIVIDUALS.USING.THE.INTERNET.2011','ISO.3166.1.alpha.2.code'))
Something like this, for example:
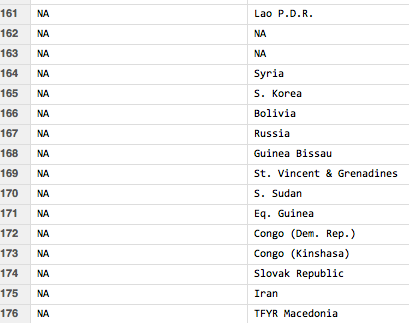
Unfortunately, not every country in the original data set is matched:
In particular, here are the unmatched items and what they presumably should have been matched with:
Lao P.D.R. - LAO PEOPLE'S DEMOCRATIC REPUBLIC Syria - SYRIAN ARAB REPUBLIC S. Korea - KOREA, REPUBLIC OF Bolivia - BOLIVIA, PLURINATIONAL STATE OF Russia - RUSSIAN FEDERATION Guinea Bissau - GUINEA-BISSAU St. Vincent & Grenadines - SAINT VINCENT AND THE GRENADINES S. Sudan - SOUTH SUDAN Eq. Guinea - EQUATORIAL GUINEA Congo (Dem. Rep.) - CONGO, THE DEMOCRATIC REPUBLIC OF THE Congo (Kinshasa) - CONGO Slovak Republic - SLOVAKIA Iran - IRAN, ISLAMIC REPUBLIC OF TFYR Macedonia - MACEDONIA, THE FORMER YUGOSLAV REPUBLIC OF
We could try to increase the Levenshtein Distance within which a partial match is suggested, but then we run the risk of introducing false positives.
Removing stopwords as part of the signature function may help, for example in the case of St. Vincent & Grenadines. (Also, I’m not sure what went wrong with Guinea Bissau?) We could presumably also make something of the abbreviations (S., Dem., Rep., Eq., P. D. R.), using these as NGrams or stubs in a regular expression? So for example, convert Lao P. D. R. to a regular expression invoking searches for a three word phrase in which consecutive words start with P, D, R. (Of course, this would require matching across the full country name rather than the signature.)
The Google Refine documentation page Clustering In Depth (Methods and theory behind the clustering functionality in Google Refine) also identifies several other fuzzy/partial matching algorithms that could be tried here. In which case, I guess I need to rewrite the above function so that it can accommodate different matching algorithms? IF you manage to implement any of these other fuzzy matching approaches, please post a link in the comments.
PS If you use any other approaches for partial matching of strings, please feel free to share how you do it in the comments below…:-) For example:
– theBioBucket: Merging Dataframes by Partly Matching String. This post also introduces adist(), an R function that returns the Levenshtein distance between two strings.

The countrycode package is quite good at handling these special cases.
@Chris – thanks; can you give an example? I have to admit, I didn’t actually look for country coding methods, I was more focussed on the general problem of partial/fuzzy matching…
countrycode gets all but one of your original names right:
> data = read.csv(url(‘http://s.telegraph.co.uk/graphics/conrad/PercentageUsingTheNet.csv’, encoding=’MACROMAN’))
> names = countrycode(data$ECONOMY, ‘country.name’, ‘country.name’)
> data$ECONOMY[is.na(names)]
[1] S. Tom� & Principe
Hi Kent — please expand on ‘countrycode’ .. that seems to do the job?
The countrycode() function is from the countrycode package suggested by Chris.
Hi Kent – out of interest, how did you know to add: encoding=’MACROMAN’ ?
@Tony – Trial and error in Chrome. I would not have noticed the need at all except the failing case had an invalid character. So I opened the CSV file in Chrome and tried different encodings until I found one that displayed correctly. MacRoman was actually my fourth choice after ISO-8859-1 (Latin-1), UTF-8 and Windows 1252.
Ah – trial and error… encodings often get me; I keep thinking there has to be a sensible way of trying to identify encodings programmatically?
Not a perfect solution yet – but I recently noticed the http://nomenklatura.okfnlabs.org/ tool, which could be used to build a public list of all possible known names of a country, and which would then provide a web service for this sort of reconciliation.
I’ve used Geonames in the past for similar tasks, but it can get mixed up with the Congos and Sudans etc.
That algorithm is quadratic (ie O(n^2)) in time, so is most suited to small lists. You can get around this by using blocking like Refine does, but then you won’t find near matches which end up in different blocks.
For the specific country code problem, simply reconciling the list against Freebase in Refine and then using “Add column from Freebase” with the “ISO Alpha 2” property will get you all these codes with no programming required. This also has the advantage that it will not only find close lexical matches, but alternative names which are completely different because Freebase typically has these alternative names as aliases.
@Tom Re O(n^2) – yes, I did ponder that; I’m not sure how efficient R’s merge is. One algorithm I’ve often pondered is a match that strips elements of the second list as they are matched, so the second list gets shorter as matches are found. I guess I need to read “Mining of Massive Datasets” again, and actually to try use some of the algorithms it describes so I can properly start to get a handle on anything other than small datasets.
As to reconciling the country code data using Freebabse: yep, agreed; but that wasn’t the focus of this post… (I did consider doing the post with random strings, but then I thought the country matching context might help folk see why I was bothering ;-)
I haven’t looked closely, but I would expect a naive matching algorithm to be O(n*m) where n and m are the lengths of the two lists. Since the list of country codes is not growing much, the matching time will probably grow linearly with the length of the list being matched.
@kent thanks for raising that; wrt n^2, I was assuming similar length lists, but of course this will not generally be the case.
Maybe I have found two bugs:
#Bring the original text into the partial match list based on the sig key.
-todo=merge(todo,yy,by.x=’partials’,by.y=’sig’)
+todo=merge(todo,yy,by.x=’partials’,by.y=’sig’,all.x=T)
#Label these rows as partial match items
-partial.matched$pass=”Partial”
+if (nrow(partial.matched) > 0) partial.matched$pass=”Partial”
Thanks for the code!
Thanks – will check those out…
Reblogged this on Manipulate Magazine: Math 4 You By Us Group Illinois.