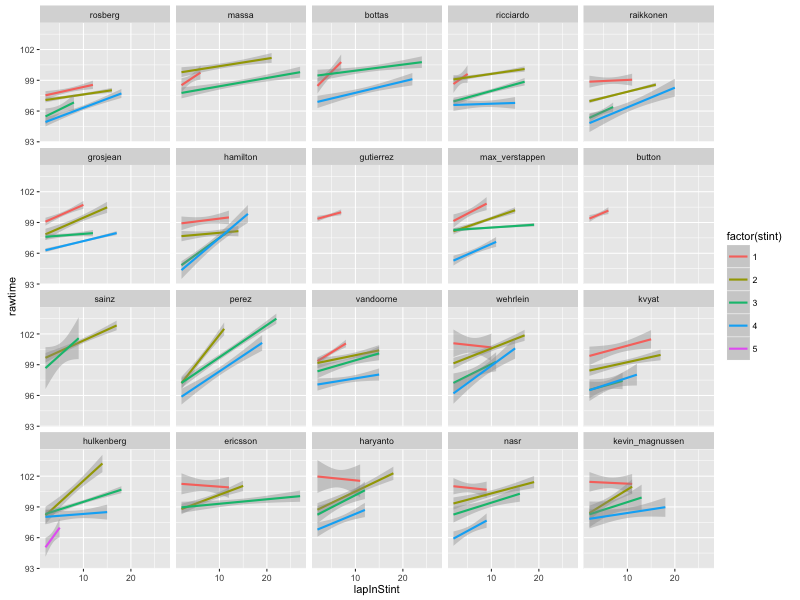
In a course team accessibility briefing last week, Richard Walker briefly mentioned a tool for automatically generating text descriptions of Statistics Canada charts to support accessibility. On further probing, the tool, created by Leo Ferres, turned out to be called iGraph-Lite:
… an extensible system that generates natural language descriptions of statistical graphs, particularly those created for Statistics Canada online publication, “The Daily”. iGraph-Lite reads a graph from a set of graphing programs, builds intermediate representations of it in several formats (XML, CouchDB and OWL) and outputs a description using NLG algorithms over a “string template” mechanism.
The tool is a C# application compiled as a Windows application, with the code available (unchanged now for several years) on Github.
The iGraph-Lite testing page gives a wide range of example outputs:

Increasingly, online charts are labeled with tooltip data that allows a mouseover or tab action to pop-up or play out text or audio descriptions of particular elements of the chart. A variety of “good chart design” principles designed to support clearer, more informative graphics (for example, sorting categorical variable bars in a bar chart by value rather than using an otherwise arbitrary alphabetic ordering) not only improves the quality of the graphic, it also makes a tabbed through audio description of the chart more useful. For more tips on writing clear chart and table descriptions, see Almost Everything You Wanted to Know About Making Tables and Figures.
The descriptions are quite formulaic, and to a certain extent represent a literal reading of the chart, along with some elements of interpretation and feature detection/description.
Here are a couple of examples – first, for a line chart:
This is a line graph. The title of the chart is “New motor vehicle sales surge in January”. There are in total 36 categories in the horizontal axis. The vertical axis starts at 110.0 and ends at 160.0, with ticks every 5.0 points. There are 2 series in this graph. The vertical axis is Note: The last few points could be subject to revisions when more data are added. This is indicated by the dashed line.. The units of the horizontal axis are months by year, ranging from February, 2005 to January, 2007. The title of series 1 is “Seasonally adjusted” and it is a line series. The minimum value is 126.047 occuring in September, 2005. The maximum value is 153.231 occuring in January, 2007. The title of series 2 is “Trend” and it is a line series. The minimum value is 133.88 occuring in February, 2005. The maximum value is 146.989 occuring in January, 2007.
And then for a bar chart:
This is a vertical bar graph, so categories are on the horizontal axis and values on the vertical axis. The title of the chart is “Growth in operating revenue slowed in 2005 and 2006”. There are in total 6 categories in the horizontal axis. The vertical axis starts at 0.0 and ends at 15.0, with ticks every 5.0 points. There is only one series in this graph. The vertical axis is % annual change. The units of the horizontal axis are years, ranging from 2001 to 2006. The title of series 1 is “Wholesale” and it is a series of bars. The minimum value is 2.1 occuring in 2001. The maximum value is 9.1 occuring in 2004.
One of the things I’ve pondered before was the question of textualising charts in R using a Grammar of Graphics approach such as that implemented by ggplot2. As well as literal textualisation of grammatical components, a small amount of feature detection or analysis could also be used to pull out some meaningful points. (It also occurs to me that the same feature detection elements could also then be used to drive a graphical highlighting layer back in the visual plane.)
Prompted by my quick look at the iGraph documentation, I idly wondered how easy it would be to pick apart an R ggplot2 chart object and generate a textual description of various parts of it.
A quick search around suggested two promising sources of structured data associated with the chart objects directly: firstly, we can interrogate the object return from calling ggplot() and it’s associated functions directly; and ggplot_build(g), which allows us to interrogate a construction generated from that object.
Here’s an example from the quickest of plays around this idea:
library(ggplot2)
g=ggplot(economics_long, aes(date, value01, colour = variable))
g = g + geom_line() + ggtitle('dummy title')
#The label values may not be the actual axis limits
txt=paste('The chart titled"', g$labels$title,'";',
'with x-axis', g$labels$x,'labeled from',
ggplot_build(g)$panel$ranges[[1]]$x.labels[1], 'to',
tail(ggplot_build(g)$panel$ranges[[1]]$x.labels, n=1),
'and y-axis', g$labels$y,' labeled from',
ggplot_build(g)$panel$ranges[[1]]$y.labels[1], 'to',
tail(ggplot_build(g)$panel$ranges[[1]]$y.labels,n=1), sep=' ')
if (<span class="pl-s"><span class="pl-pds">'</span>factor<span class="pl-pds">'</span></span> <span class="pl-k">%in%</span> class(<span class="pl-smi">x</span><span class="pl-k">$</span><span class="pl-smi">data</span>[[<span class="pl-smi">x</span><span class="pl-k">$</span><span class="pl-smi">labels</span><span class="pl-k">$</span><span class="pl-smi">colour</span>]])){
txt=paste(txt, '\nColour is used to represent', g$labels$colour)
if ( class(g$data[[g$labels$colour]]) =='factor') {
txt=paste(txt,', a factor with levels: ',
paste(levels(g$data[[g$labels$colour]]), collapse=', '), '.', sep='')
}
}
txt
#The chart titled "dummy title" with x-axis date labeled from 1970 to 2010 and y-axis value01 labeled from 0.00 to 1.00
#Colour is used to represent variable, a factor with levels: pce, pop, psavert, uempmed, unemploy.&amp;amp;quot;
So for a five minute hack, I’d rate that approach as plausible, perhaps?!
I guess an alternative approach might also be to add an additional textualise=True property to the ggplot() call that forces each ggplot component to return a text description of its actions, and/or features that might be visually obvious from the action of the component as well as, or instead of, the graphical elements. Hmm… maybe I should use the same syntax as ggplot2 and try to piece together something along the lines of a ggdescribe library? Rather than returning graphical bits, each function would return appropriate text elements to the final overall description?!
I’m not sure if we could call the latter approach a “transliteration” of a chart from a graphical to a textual rendering of its structure and some salient parts of a basic (summary) interpretation of it, but it feels to me that this has some merit as a strategy for thinking about data2text conversions in general. Related to this, I note that Narrative Science have teamed up with Microsoft Power BI to produce a “Narratives for Power BI” plugin. I imagine this will start trickling down to Excel at some point, though I’ve already commented on how simple text summarisers can be added into to Excel using simple hand-crafted formulas (Using Spreadsheets That Generate Textual Summaries of Data – HSCIC).
PS does anyone know if Python/matplotib chart elements also be picked apart in a similar way to ggplot objects? [Seems as if they can: example via Ben Russert.]
PPS In passing, here’s something that does look like it could be handy for simple natural language processing tasks in Python: polyglot, “a natural language pipeline that supports massive multilingual applications”; includes language detection, tokenisation, named entity extraction, part of speech tagging, transliteration, polarity (i.e. crude sentiment).
PPPS the above function is now included in Jonathan Godfrey’s BrailleR package (about).
PPPPS for examples of best practice chart description, see the UK Association for Accessible Formats (UKAAF) guidance on Accessible Images.
PPPPPS rules for converting eg MathJax and MathML to speech, braille, etc
PPPPPPS a possibly handy R package — equatiomatic — for generating a LaTeX equation that describes a fitted R model. In Python, erwanp/pytexit will “convert a Python expression to a LaTeX formula”.